Organising research data into the correct format can be time consuming, existing software may not be optimised for your data or you may have no way of of automating repetitive processes.
We are able to create custom software for use on work stations, laptops, servers, high-performance computing clusters or even mobile devices.
Our engineers are fluent in multiple programming languages and up-to-date with cutting edge algorithmic and graphical techniques. With years of industry experience the team work together professionally and are motivated to help you get the most from your experiments and to ensure your findings are accessible to the world.
Automation
We can help you understand complex workflows and improve them with automation.
Dr Sara Silva Pereira commissioned CBF to create an efficient python pipeline of her genomic, transcriptomic and HMM methods used to produce Antigen Variant Profiles of Trypanosome spp. Multiple methods and species were combined into one python program and the tool was published for the Galaxy. An application note is under review at Bioinformatics.
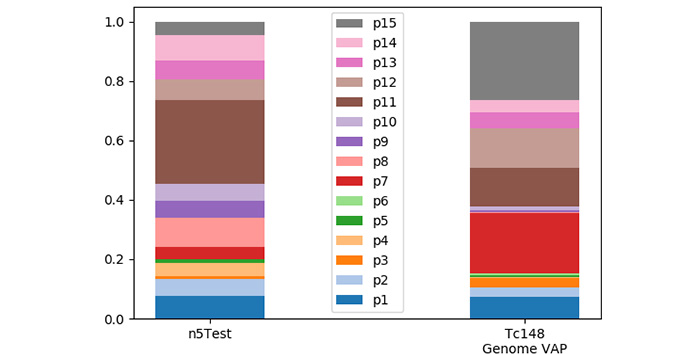
The transcriptomic Variant Antigen Profile of Trypanosoma congolense estimated as phylotype proportion adjusted for transcript abundance and the reference genomic Variant Antigen Profile. Data was produced with the ‘Variant Antigen Profiler’ (Silva Pereira and Jackson, 2018)
Sharing methods while retaining control
If you want to share your methods for others to use but still want to retain control of the method itself, we can help.
Dr Jenny Hodgson developed the high-impact research project condatis. A decision support tool to identify the best locations for habitat creation and restoration to enhance networks and increase connectivity across landscapes.
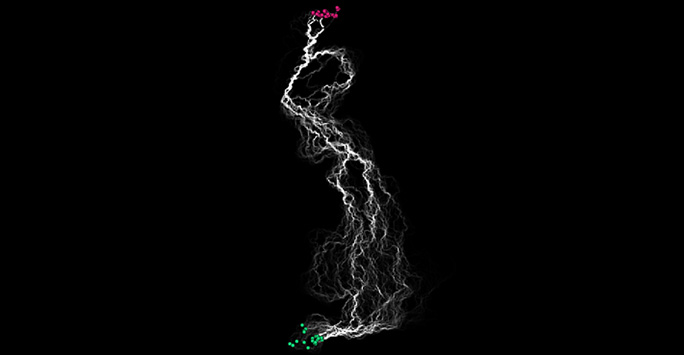
British Grasslands – 2km2 cells. Connectivity pathways from source (green) to target (red) Modelled in condatis v0.6
Databases
We can help formalise your datasets into a readily available database online.
We helped create the Allele Frequency website that does just that. The Allele Frequency net database has more than 10000 unique users per year, from across disciplines including transplant immunology, vaccine development, pharmacogenetics, human genetics and anthropology and infectious diseases.
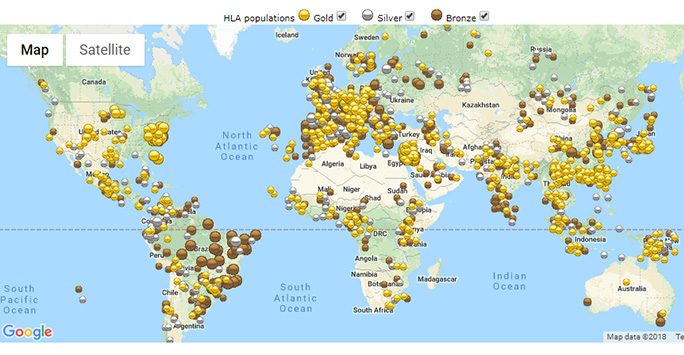
HLA populations in the Allele Frequency Net Database (by data quality standard)
Data visualisation
We can create a novel and interactive visualisation of your data.
For example Channel Bank the online platform for testing and developing deep learning analysis of ion channels.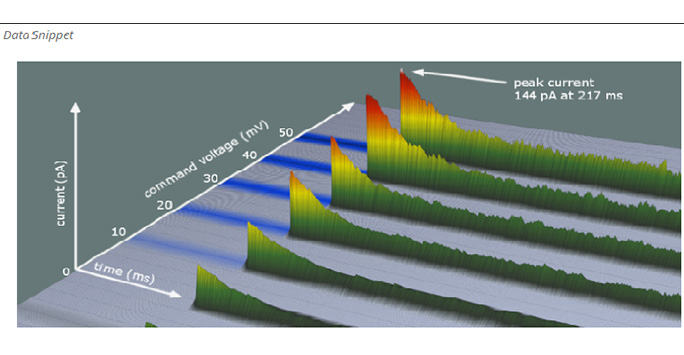
Data visualisation
IcmsWorld
Dr Tony McCabe has developed software which is a high-performance tool for efficient 3D visualisation of LC-MS data. You can use it to quickly load and view (large) .mzml and .raw files, for both proteomic and metabolomics experiments. You can view the whole file at once, then smoothly zoom in to view any areas of interest in full detail, down to each individual data point.
An identification file can be loaded; these identifications can be overlaid onto the LC-MS data, filtered, and selected to zoom directly to the relevant raw data.
Multiple files can be viewed simultaneously, side-by-side, to make it easy to compare data sets. By setting up the files on a webserver, datasets can be viewed remotely with lcmsWorld, without having to download the entire file.
If you would like to test this software please contact Dr McCabe.
Back to: Computational Biology Facility