The team covers protein modelling of all kinds and the use of such models for applications, from biomedical eg structure-based SNP interpretation, to biotechnological eg genome mining and protein design.
Protein redesign for commercially valuable enzyme activities
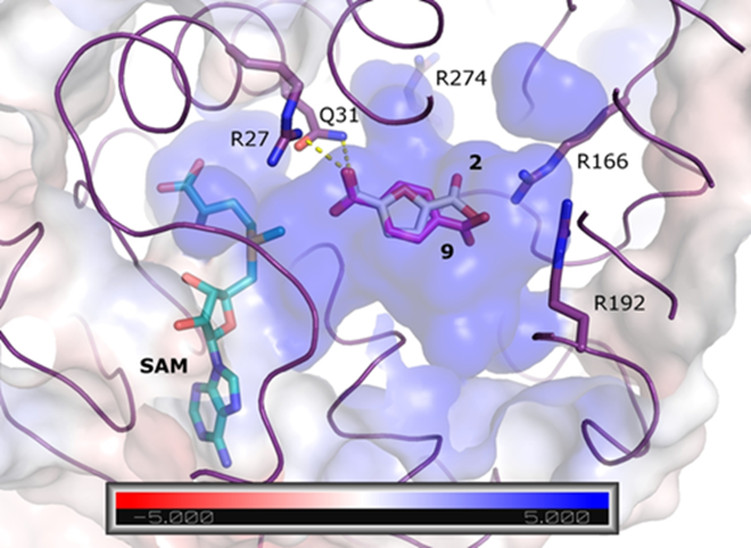
Synthesis of bioplastics from cellulose waste offers a route to renewable replace current polymers with biodegradable alternatives. In collaboration with Dr Andrew Carnell (Department of Chemistry) structure-based analyses of AlphaFold 2 models and small molecule docking were used to suggest mutations to reshape the natural catalytic site of a fungal protein, successfully enhancing activity. The work has been published here and here.
Cofolding with AlphaFold 2 elucidates the structure of bacterial functional amyloid fibrils
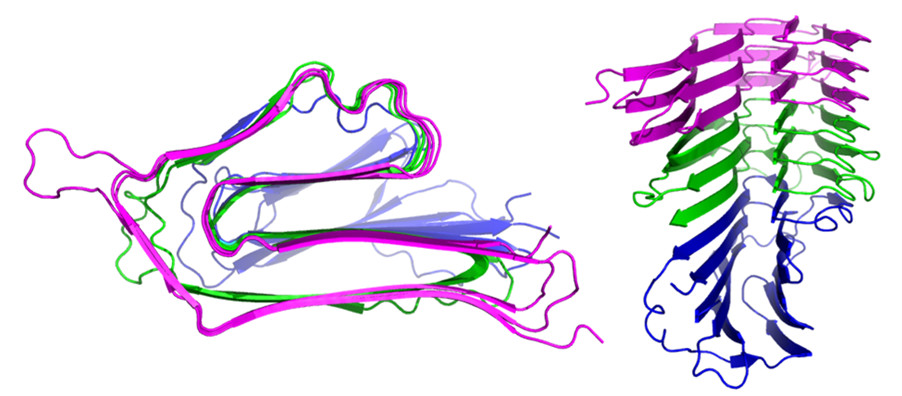
In collaboration with Jill Madine, modelling of the complex between three bacterial proteins was done. It shows how two, in magenta and green, share a novel amyloid architecture, while a regular globular protein in blue caps the fibril.
Function annotation of a human membrane transporter
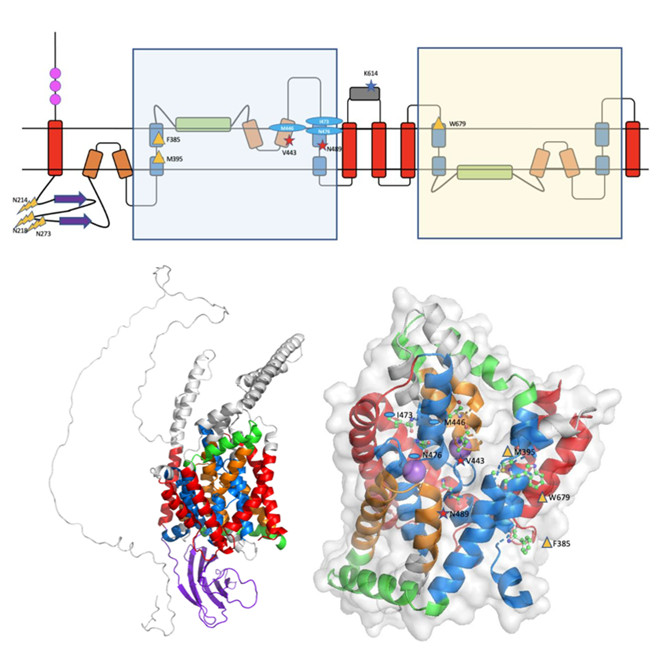
Assigning function to cryptic proteins is of major importance. Work by Shah Mesdaghi, now employed in the CBF, used AlphaFold 2 and structure-based function annotation methods to address the Oca2 protein associated with human albinism. It revealed an unsuspected GOLD domain in purple, linked to subcellular transport; overturned the prevailing view of its topology; and enabled mapping of known damaging mutations.
Putting Shigella mutations and epitopes in a structural context
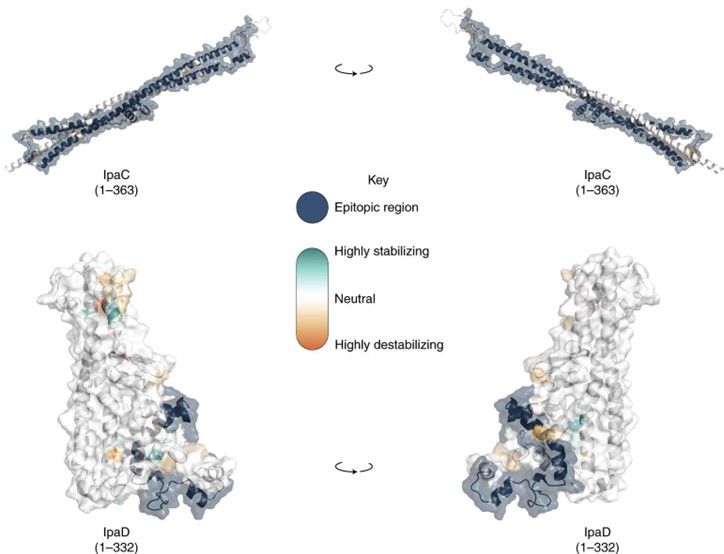
In collaboration with Kate Baker, AlphaFold 2 modelling of several vaccine protein antigens was done. This allowed mutations of differing severity to be placed within or outside of known epitopes with implications for future pathogen monitoring and therapy. The work was published here.
Determining the dynamics properties of an enzyme
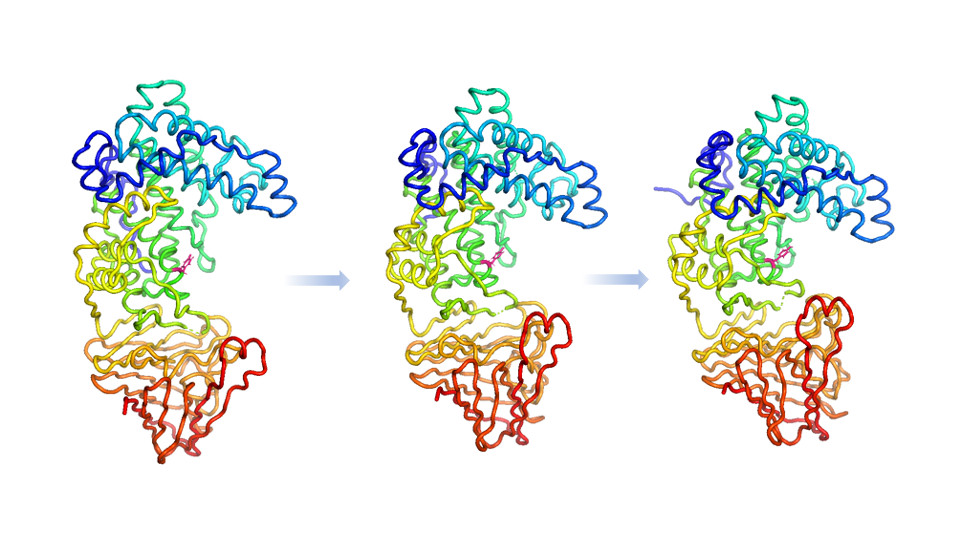
Especially post-AlphaFold 2, consideration of the dynamic properties of macromolecules is very important. In collaboration with Alan Cartmell, Molecular Dynamics simulations of a new crystal structure were carried out. This allowed the prediction of how domain motions bring together residues forming the catalytic site.
Back to: Computational Biology Facility