Networks lie at the heart of many applications in modern science. From social networks to cell signalling, networks provide insights about the organisation of complex systems.
We can use data from your experiments and combine it with many public domain resources for network generation.
The case studies below demonstrate the applications and flexibility of network-based systems biology approaches:
Mapping of molecular signatures onto protein-protein interaction networks
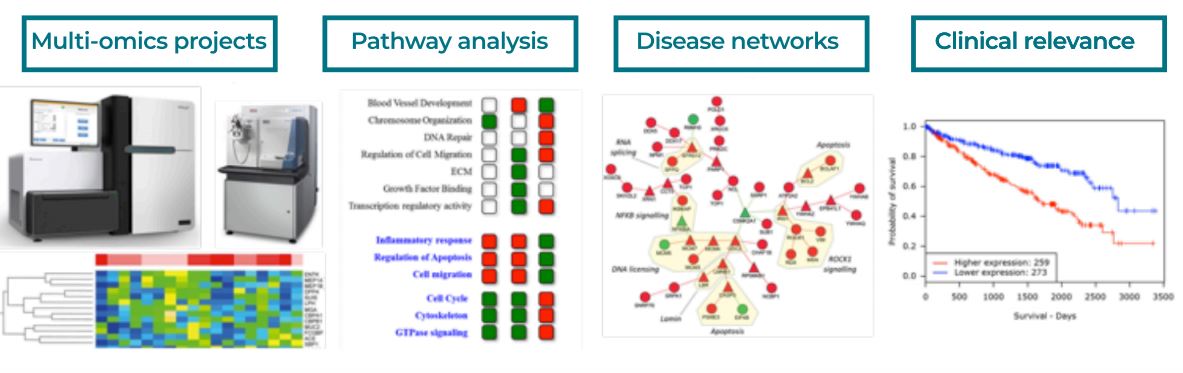
- This study explored the role of the extracellular matrix protein Thrombospondin-1 (THBS1) in glioblastoma (GBM) development
- We show that the TGFβ canonical pathway transcriptionally regulates THBS1, through SMAD3 binding to the THBS1 gene promoter. THBS1 silencing inhibits tumour cell invasion and growth, alone and in combination with anti-angiogenic therapy.
- RNA sequencing of patient-derived xenograft tissue from laser capture micro-dissected peripheral and central tumour areas identified signatures of angiogenic and invasive portions of the tumour.
- Using an integrative approach, we constructed protein-protein interaction networks representing potential signalling events between tumour and stroma cells. Briefly, this consisted of integraitng:
- the expression signature of invasive tumour areas
- genes expressed in the mouse stroma surrounding invasive tumour cells
- map the expressions above onto a protein-protein interaction scaffold representing experimentally validated interactions from the BioGrid database
- We could show that THBS1 is one of the genes with the highest connectivity at these tumour borders
- The gene expression profiling and network analysis provided a highly complementary systems level view of the invasive areas of the tumour, the management of which represents a significant challenge in the treatment of GBM.
Read the full journal article, 'Deciphering the complex role of thrombospondin-1 in glioblastoma development'.
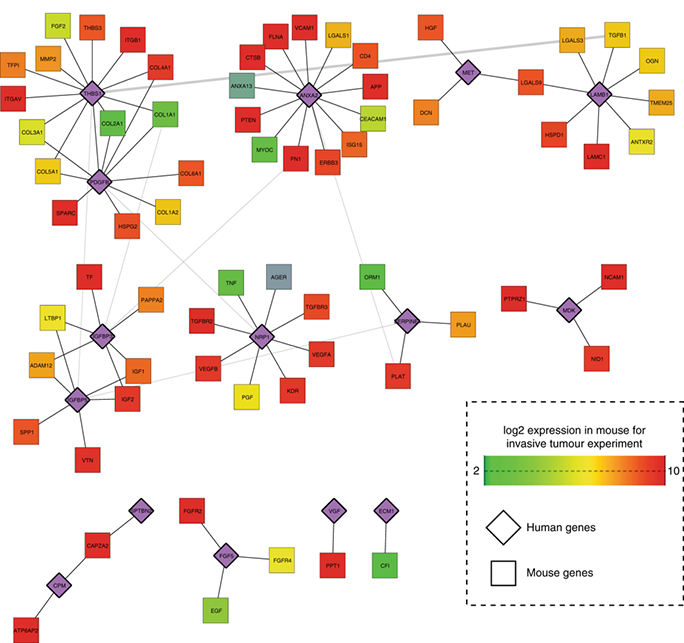
Figure 1: Transcriptional analysis highlights THBS1 as central regulator of GBM invasion. A network of proteins encoded by genes that are overexpressed in the invasive area of P3 tumours when compared to the core area, or are expressed by the host. Relationships in the network represent known protein–protein interactions. Human genes are represented in purple and mouse genes are colour coded according to their expression (green (2) – red (10); normalised counts in log2 scale).
Clarke et al., Cell Rep, 2017
- Co-expression networks representing the effect of exercise on human muscle revealed a role of Eif6 in controlling metabolic adaptation
- For this work Clarke et al. integrated functional genomics data, information regarding known biological pathways relevant in human muscle, and clinical data collected from human participants during the study (see figure 2)
- The network analysis provided robust hypotheses that were experimentally validated using a mouse knock-out model.
Figure 2: A biological network representing the integration of gene expression, pathway and clinical data that allowed us to generate hypotheses to explain the linkage between a single molecule of interest, regulation of downstream processes and phenotypic variation.
Clarke and Daubon et al., PLoS Genet, 2015
- A novel network searching algorithm was applied to integrate multiple functional genomics data and reveal a potential mechanism explaining the pro-tumour activity of Rho GTPases in cancer
- For this work the authors used integration of multiple layers of data including large patient datasets available in the public domain and in-house proteomics data (see figure 3)
- Results were then validated using cell models and imaging approaches.
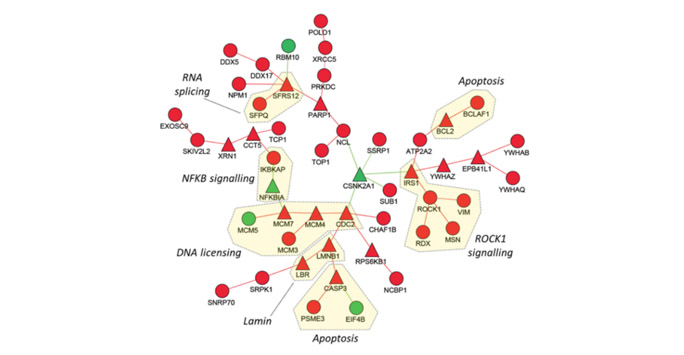
Figure 3: A network biology approach predicts a new mechanism of a pro-tumour gene. By integrating novel protein interaction data with functional genomics data from patients we can predict a new mechanism for a pro-tumour gene. Using the network model above we predicted that RND3 silencing led to a disruption of the cell cycle caused by defective DNA licensing complexes.
Back to: Computational Biology Facility