Vicky Harman 2012-current Proteome changes during protein overexpression
Victoria and Rob are working on Rob’s fellowship project. We have a number of ongoing projects, centering on global proteomics changes and novel methods to simplify the proteome prior to proteomic analysis.
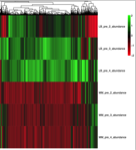
During the COPY project, a global absolute quantification of the yeast proteome using QconCAT technology, we recombinantly expressed over 100 QconCAT protiens. We found that some QconCAT proteins failed to express, or did not express to detectable levels. Our interest was piqued and we determined to investigate whether there were global proteomic changes to be seen in E.coli following the addition of the QconCAT plasmid, or the addition of Isopropyl β-D-1-thiogalactopyranoside (IPTG) the synthetic analog of lactose used to induce expression of recombinant proteins using the pET21a vector system. Our study starts by looking at the large scale proteomic changes that occur when E.coli is grown in different types of media, and whether E.coli, without a pET21a plasmid vector, is influenced by the presence of IPTG. We hope to progress this work on to look at the effect of the presence of a plasmid, containing either an expressing or non-expressing QconCAT.
Victoria and Rob are working on Rob’s fellowship project. We have a number of ongoing projects, centering on global proteomics changes and novel methods to simplify the proteome prior to proteomic analysis.
During the COPY project, a global absolute quantification of the yeast proteome using QconCAT technology, we recombinantly expressed over 100 QconCAT protiens. We found that some QconCAT proteins failed to express, or did not express to detectable levels. Our interest was piqued and we determined to investigate whether there were global proteomic changes to be seen in E.coli following the addition of the QconCAT plasmid, or the addition of Isopropyl β-D-1-thiogalactopyranoside (IPTG) the synthetic analog of lactose used to induce expression of recombinant proteins using the pET21a vector system. Our study starts by looking at the large scale proteomic changes that occur when E.coli is grown in different types of media, and whether E.coli, without a pET21a plasmid vector, is influenced by the presence of IPTG. We hope to progress this work on to look at the effect of the presence of a plasmid, containing either an expressing or non-expressing QconCAT.
Sam Ferries 2013-2017 (title)
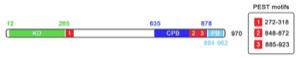
My PhD project is focused on the serine/threonine kinase, polo-like kinase 4 (PLK4), a key regulator of centrosome biogenesis which ensures centrioles are duplicated once per cell cycle, allowing the normal segregation of chromosomes prior to cytokinesis. Both over and under-expression of PLK4 can lead to over-duplication of centrioles, leading to aberrant chromosome segregation, an important hallmark of many human cancers. Despite its importance, the signalling pathway in which PLK4 operates is poorly understood, with very few substrates having been identified. My PhD project aims to advance the understanding of the role of PLK4 using chemical genetics and a mass spectrometry based approach to discover substrates and uncover the signalling network in which it operates, potentially discovering novel biomarkers for certain human cancers.
Drug resistant mutants will be generated alongside the wild type protein and stably transfected in to a human cancer cell line. Both cell lines will be screened with a panel of inhibitors and, by SILAC followed by phosphopeptide enrichment and LC-MS/MS, potential substrates can be identified and quantified using the drug resistant mutant as a tool to validate results due to the promiscuous nature of kinase inhibitors. This allows identified phosphpeptides to be attributed to PLK4 and will begin to unravel the PLK4 regulated phosphoproteome.
My PhD project is focused on the serine/threonine kinase, polo-like kinase 4 (PLK4), a key regulator of centrosome biogenesis which ensures centrioles are duplicated once per cell cycle, allowing the normal segregation of chromosomes prior to cytokinesis. Both over and under-expression of PLK4 can lead to over-duplication of centrioles, leading to aberrant chromosome segregation, an important hallmark of many human cancers. Despite its importance, the signalling pathway in which PLK4 operates is poorly understood, with very few substrates having been identified. My PhD project aims to advance the understanding of the role of PLK4 using chemical genetics and a mass spectrometry based approach to discover substrates and uncover the signalling network in which it operates, potentially discovering novel biomarkers for certain human cancers.
Drug resistant mutants will be generated alongside the wild type protein and stably transfected in to a human cancer cell line. Both cell lines will be screened with a panel of inhibitors and, by SILAC followed by phosphopeptide enrichment and LC-MS/MS, potential substrates can be identified and quantified using the drug resistant mutant as a tool to validate results due to the promiscuous nature of kinase inhibitors. This allows identified phosphpeptides to be attributed to PLK4 and will begin to unravel the PLK4 regulated phosphoproteome.
Richard Bennett 2013-2016 (title)
My PhD project is exploring strategies for absolute protein quantification. This is becoming increasingly important in the field of proteomics, owing to the significance the resultant data plays in, for example, systems biology and biomarker development. Most mass spectrometry (MS) based quantification experiments employ a stable isotope labelled internal standard to quantify proteolytic peptides unique to the protein of interest. Internal standards can take the form of full length recombinant proteins, synthesised peptides or artificial proteins consisting of concatamers of the so-called quantotypic peptides (QconCATs). These standards are used in conjunction with selected reaction monitoring (SRM), in which only specific precursor/product ion pairs are chosen for detection, to create an exquisitely sensitive targeted assay for quantification.
As well as MS, western blotting (WB) can also be used for quantification. This relies on antibodies to specifically detect a target in a complex sample. Endogenous proteins can be quantified by means of a calibration curve created using a purified protein standard. WB has the advantage of being more sensitive than MS, however it lacks the specificity of SRM, as well as being notoriously laborious, unreproducible and low throughput. To overcome some of the shortfalls of western blotting and improve data quality, automated capillary western technology from Protein Simple is being used for quantitative WB experiments.
My PhD project is exploring strategies for absolute protein quantification. This is becoming increasingly important in the field of proteomics, owing to the significance the resultant data plays in, for example, systems biology and biomarker development. Most mass spectrometry (MS) based quantification experiments employ a stable isotope labelled internal standard to quantify proteolytic peptides unique to the protein of interest. Internal standards can take the form of full length recombinant proteins, synthesised peptides or artificial proteins consisting of concatamers of the so-called quantotypic peptides (QconCATs). These standards are used in conjunction with selected reaction monitoring (SRM), in which only specific precursor/product ion pairs are chosen for detection, to create an exquisitely sensitive targeted assay for quantification.
As well as MS, western blotting (WB) can also be used for quantification. This relies on antibodies to specifically detect a target in a complex sample. Endogenous proteins can be quantified by means of a calibration curve created using a purified protein standard. WB has the advantage of being more sensitive than MS, however it lacks the specificity of SRM, as well as being notoriously laborious, unreproducible and low throughput. To overcome some of the shortfalls of western blotting and improve data quality, automated capillary western technology from Protein Simple is being used for quantitative WB experiments.