Molecular Insights into Parkinson’s Disease: Deciphering the Mechanism of Parkin E3 Ligase and PINK1 Kinase
- Sonia Rocha
- Suitable for: Staff and students with an interest in Genomes, Systems and Therapeutic Targeting
- Admission: Free
Add this event to my calendar
Click on "Create a calendar file" and your browser will download a .ics file for this event.
Microsoft Outlook: Download the file, double-click it to open it in Outlook, then click on "Save & Close" to save it to your calendar. If that doesn't work go into Outlook, click on the File tab, then on Open & Export, then Open Calendar. Select your .ics file then click on "Save & Close".
Google Calendar: download the file, then go into your calendar. On the left where it says "Other calendars" click on the arrow icon and then click on Import calendar. Click on Browse and select the .ics file, then click on Import.
Apple Calendar: The file may open automatically with an option to save it to your calendar. If not, download the file, then you can either drag it to Calendar or import the file by going to File >Import > Import and choosing the .ics file.
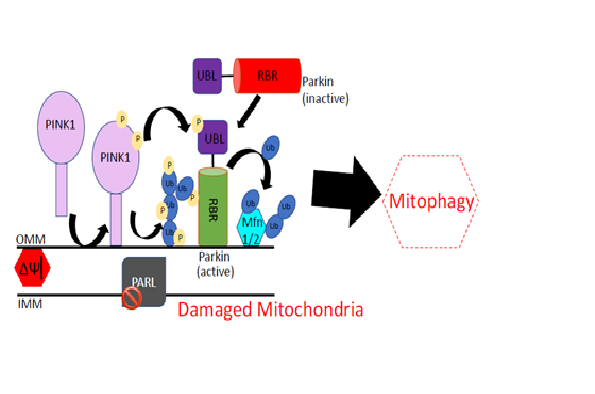
Parkinson’s disease (PD) is a neurodegenerative disorder with severe motor and non-motor symptoms. Mutations on several genes associated with various structural and functional components of neurons cause loss of neurons which results in PD. However, two genes PARK2 and PARK6 are found most frequently mutated causing more than 50% of the familial form of the disease. PARK2 gene encodes an E3 ubiquitin ligase Parkin, and PARK6 encodes PTEN-induced Kinase 1 (PINK1). Parkin is an auto-inhibited enzyme mediated by its ubiquitin like (UBL) domain. During stress condition, PINK1 is stabilised on mitochondria and phosphorylates Ser65 on UBL domain of Parkin and ubiquitin. Phosphorylation of Parkin and ubiquitin results in fully active Parkin which clears the damaged mitochondria, however, underlying molecular mechanism of PINK1 and Parkin signalling is poorly understood. I have determined human Parkin structures in both in-active state (apo-Parkin) & active state (phospho-mimic Parkin & in complex with phospho-ubiquitin (pUb)). My research on Parkin reveals how Ubl domain cause inhibition, and how Parkin is allosterically activated by pUb binding. My research reveals an intriguing mechanism of catalysis of Parkin E3 ubiquitin ligase. I have also solved the first crystal structure of PINK1 and identified a unique insertion in the kinase domain responsible for specific recognition of ubiquitin/UBL domain of Parkin. Mapping of mutations on PINK1 and Parkin structures suggests that mutations are present in important functional regions perturbing function of these enzymes and cause Parkinson’s. The study provides molecular rationale for therapeutics purpose, and lays foundation of future research towards identifying key enzymes involved in phospho-ubiquitin signalling along with establishing the role of other genes associated with the disease.
