The Drosophila-Spiroplasma Interaction as a Model for Endosymbiosis Genetics
- Ewa Chrostek
- Suitable for: Staff and students with an interest in Behaviour, Evolution, Ecology and Microbiology
- Admission: Free
Add this event to my calendar
Click on "Create a calendar file" and your browser will download a .ics file for this event.
Microsoft Outlook: Download the file, double-click it to open it in Outlook, then click on "Save & Close" to save it to your calendar. If that doesn't work go into Outlook, click on the File tab, then on Open & Export, then Open Calendar. Select your .ics file then click on "Save & Close".
Google Calendar: download the file, then go into your calendar. On the left where it says "Other calendars" click on the arrow icon and then click on Import calendar. Click on Browse and select the .ics file, then click on Import.
Apple Calendar: The file may open automatically with an option to save it to your calendar. If not, download the file, then you can either drag it to Calendar or import the file by going to File >Import > Import and choosing the .ics file.
Insects frequently maintain symbiotic relationships with vertically transmitted bacterial partners that live within their body called endosymbionts. Endosymbiotic infections have major impacts on insects affecting their nutrition, their reproduction and their ability to resist environmental challenges. Deciphering the molecular dialogue that underlies host-endosymbiont interactions is thus of major importance to better understand the physiology and evolution of insects. However, functional research on insect endosymbiosis is often hampered by the lack of robust tools that allow genetic manipulation of both the host and the symbionts.
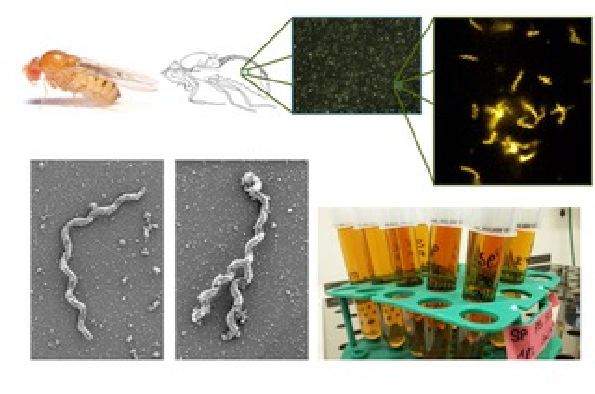
Spiroplasma are widespread arthropod-associated bacteria that are helical, highly mobile, and do not possess any cell wall. S. poulsonii is an endosymbiotic species that infects Drosophila melanogaster. This is an ideal model to study the molecular basis of insect endosymbiosis because of its overall tractability. Using only fly genetics, we are already able to functionally characterize some Spiroplasma proteins and decipher their impact on the interaction.
Directly studying the bacterial side of endosymbiosis at large scale is however limited by the unculturability and genetic intractability of most endosymbionts. To expand our toolbox, we developed a protocol for a sustainable in vitro culture of Spiroplasma. By comparing the transcriptome of the bacterium in vitro and in the host, we identified genes potentially relevant for host-symbiont interaction. These are now priority targets for the development of a knockout method that will pave the road for in-depth functional studies on insect-bacteria symbiotic interactions with genetic tools available for both partners.
