What can we learn from Genomics and Transoposon Mutagenesis in the (Fission) Yeast Model?
- Andrea Betancourt
- Suitable for: Staff and students with an interest in Behaviour, Evolution, Ecology and Microbiology
- Admission: Free
- Book now
Add this event to my calendar
Click on "Create a calendar file" and your browser will download a .ics file for this event.
Microsoft Outlook: Download the file, double-click it to open it in Outlook, then click on "Save & Close" to save it to your calendar. If that doesn't work go into Outlook, click on the File tab, then on Open & Export, then Open Calendar. Select your .ics file then click on "Save & Close".
Google Calendar: download the file, then go into your calendar. On the left where it says "Other calendars" click on the arrow icon and then click on Import calendar. Click on Browse and select the .ics file, then click on Import.
Apple Calendar: The file may open automatically with an option to save it to your calendar. If not, download the file, then you can either drag it to Calendar or import the file by going to File >Import > Import and choosing the .ics file.
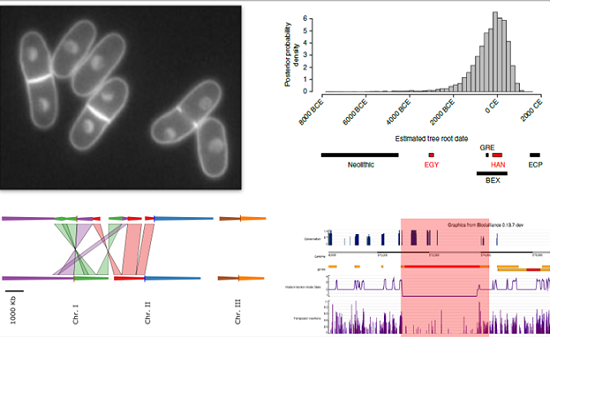
The fission yeast Schizosaccharomyces pombe is a powerful model for molecular and cellular biology. In contrast to the budding yeast, we knew very little about the diversity, ecology or evolution of the species until very recently.
I’ll introduce what little is known about the ecology of this yeast (1), and then describe three of my studies of genome diversity and function. First, I’ll outline our study of genomic and phenotypic diversity, where we describe population structure, date the dispersal of the species and show that genome-wide association studies are feasible in this species (2). I’ll then describe our analysis of structural variation from short read and long read sequencing data (3, 4) demonstrating that copy number variants both contribute to heritable traits, are unstable, and that quantitative traits are explained by an ancient admixture event from two populations.
On a slightly different track, I’ll describe our recent analysis of saturating transposon mutagenesis in S. pombe (5). In this study, we generated very dense transposon insertion libraries using the Hermes transposon, to one insertion per 13 nt of the genome. We developed a hidden Markov Model that uses the transposon insertion density to classify the relative importance of each position in the genome. We show that HMM states assign similar functional constraints to comparative genomics and genetic diversity, but with far higher resolution. This data will bring us closer to quantifying the functional significance of every base in the genome.
References
1. D. C. Jeffares, The natural diversity and ecology of fission yeast. Yeast. 35, 253–260 (2017).
2. D. C. Jeffares et al., The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nature Genetics. 47, 235–241 (2015).
3. D. C. Jeffares et al., Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 8, 14061 (2017).
4. S. Tusso et al., Ancestral admixture and structural mutation define global biodiversity in fission yeast. bioRxiv, 415091 (2018).
5. L. Grech et al., Fitness Landscape of the Fission Yeast Genome. bioRxiv, 398024 (2018).
